Published on: Nov 18, 2022
Super fast Python (Part-4): Cython
This is the fourth post in the series on Python performance and Optimization. The series points out the utilization of inbuilt libraries, low-level code conversions, and other Python implementations to speed-up Python. The other posts included in this series are
- (Part-1): Why Python is slow?
- (Part-2): Good practices to write fast Python code
- (Part-3): Multi-processing in Python
- (Part-4): Use Cython to get speed as fast as C (this post)
- (Part-5): Use Numba to speed up Python Functions and Numeric calculations
In the last post, we discussed multiprocessing to optimize Python code by utilizing parallel computing with multiple CPU cores. Multi-processing is useful when we can split certain parts of code into parallel tasks and then execute them parallelly.
Imagine when we don't have any parallelizable code and it is taking huge time in Python compared to C/C++, how to optimize the code then? One thing we can do is to convert the Python code into a low-level programming language like C/C++ and embed that C/C++ code in Python. Understanding and writing C/C++ code is hard for someone who is not familiar with C/C++. Fortunately, we can get the performance speed as fast as C/C++ by writing the Python code in Cython which is a superset of Python that provides functionality to write C-Extensions for Python.
How Cython can improve Speed?
In our previous discussion on Why Python is slow?, we learned that the major speed issues arise in Python due to
- interpretation of generated bytecode
- dynamic types and their management
With Cython, we can take care of the above problems with both compiled code instead of interpretation and static typing instead of dynamic typing.
Cython translates the Python code into C extension code and compiles the C code into an object code that can be imported directly into Python. Also, we can make some changes like adding static types that improve the execution speed drastically over dynamic typing. Cython also supports the usage of C/C++ libraries and functions that are super-fast compared to Python libraries and functions.
Install Cython
Install the Cython with Pip as follows
1pip install Cython 2
To compile the C code into an object file, we need a C compiler. Ubuntu comes with gcc by default. For other platforms like Windows, install the C compiler if not installed previously.
Cython usage
There are multiple ways to use Cython like building manually, using in Jupyter as an extension, or importing directly like a Python module without compilation using pyximport.
Build a Cython module manually
We write the Cython code in a file with the extension .pyx instead of the normal Python extension .py.
The compiler translates '.pyx' Cython file into a '.c' C file and then compiles that C file to a sharable object file '.so' (or '.pyd' on Windows). We tell the compiler those build instructions and compilation options by writing a setup.py file.
Cython compilation
- Translate the .pyx source code to a .c file with additional wrappers of Python extension code.
- Compile the .c file with a C compiler to a platform-specific shared object file .so that can be imported directly into Python.
calculate_y_cython.pyx
1def fx(x, a, b, c): 2 d = (a + b) * c 3 return a*x + b*(x**2) + c*(x**3) + d 4 5def y(x, n, a, b, c): 6 af = fx(x, a, b, c)**2 7 k = abs(n//2 - x) 8 bf = fx(k, a, b, c) 9 return (af-bf)/(n-1 + 1e-12) 10 11def calculate_y_cython(n): 12 ys = [] 13 a, b, c = 2, 5, -4 14 for i in range(n): 15 ys.append(y(i, n, a, b, c)) 16 17 return ys 18
The above file calculate_y.pyx contains Cython code that looks the same as Python without any optimizations. Now to compile the above file, write a setup.py file as following
setup.py
1from distutils.core import setup 2from Cython.Build import cythonize 3 4setup( 5 name='Calculate Expression Y', 6 ext_modules=cythonize( 7 "calculate_y_cython.pyx", 8 compiler_directives={"language_level": "3"}, 9 ) 10) 11
In the above setup.py, for the setup(), we have passed the optional name to our Cython module for name and the path to the .pyx file for the ext_module parameter.
Build the sharable object file '.so' using the following command in the terminal
1python setup.py build_ext --inplace 2or 3python setup.py build_ext --inplace --quiet 4
This will generate a translated '.c' C file and a compiled '.so' file.
We can import the above compiled calculate_y_cython module directly into Python runtime.
main.py
1from time import perf_counter 2from calculate_y_cython import calculate_y_cython 3 4def fx(x, a, b, c): 5 d = (a + b) * c 6 return a*x + b*(x**2) + c*(x**3) + d 7 8def y(x, n, a, b, c): 9 af = fx(x, a, b, c)**2 10 k = abs(n//2 - x) 11 bf = fx(k, a, b, c) 12 return (af-bf)/(n-1 + 1e-12) 13 14def calculate_y_py(n): 15 ys = [] 16 a, b, c = 2, 5, -4 17 for i in range(n): 18 ys.append(y(i, n, a, b, c)) 19 20 return ys 21 22def main(): 23 n = 100000 24 # Python implementation 25 atime = perf_counter() 26 res = calculate_y_py(n) 27 print(f'Python Time: {perf_counter()-atime:.2}') 28 29 atime = perf_counter() 30 res = calculate_y_cython(n) 31 print(f'Cython Time: {perf_counter()-atime:.2}') 32 33if __name__=="__main__": 34 main() 35 36"""Output: 37Python Time: 0.19 38Cython Time: 0.16 39""" 40
At the time of writing, the latest Python version is Python 3.11 which is incredibly faster (30-60% in some cases) than earlier versions. So, to understand the Cython potential, I'm running the scripts in Python 3.8.10 on my old system with 8GB Intel(R) i5-8250U 1.60GHz CPU on HP Laptop 15-da0xxx.
In the above main.py, we have imported the calculate_y_cython module at line 2. If we look at the output to check the time taken for normal Python implementation and Cython version, they are 0.19 and 0.16 seconds respectively. The time difference is very low and we didn't gain much from Cython because we haven't done any optimization steps like
- static typing
- limit calling Python's libraries
- reducing Python's PyObject usage
Cython Annotations
Cython provides an easy way to check where we can optimize our Cython code by using Cython annotate feature. With the following command (-a denotes annotate and -3 denotes language_level which is Python3), cython generates an HTML file that we can open in the browser to check for optimizable code. Another option is to pass the annotate=True parameter to cythonize() function call in ext_modules in setup.
1cython -a calculate_y_cython.pyx -3 2
If we open the generated HTML file in the browser, it will look like this
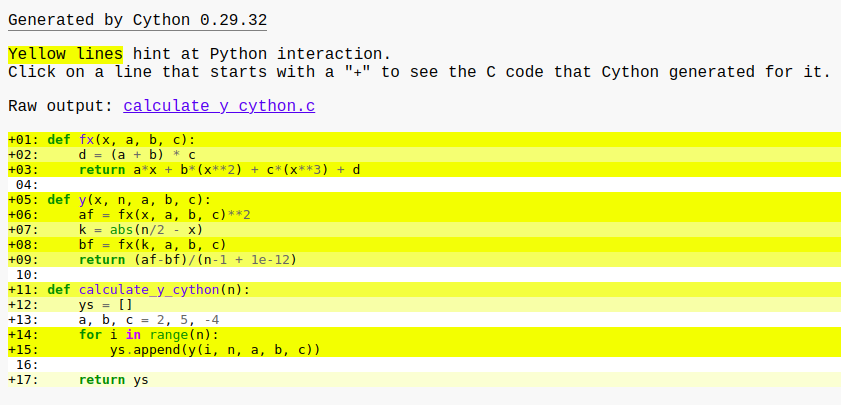
The more yellow lines the more interaction with the Python interpreter. Our goal should be converting as many yellow lines as to white lines that denote pure Cython code. We discuss the optimization part in a later section.
Cython as an extension in Jupyter
Cython can be imported and used in Jupyter directly as an extension without the need for any additional build/compilation steps.
First load the Cython extension using %load_ext cython, and then, for the cell that is to be Cythonized, use the magic command %%cython at the top of that cell as shown in the following image.
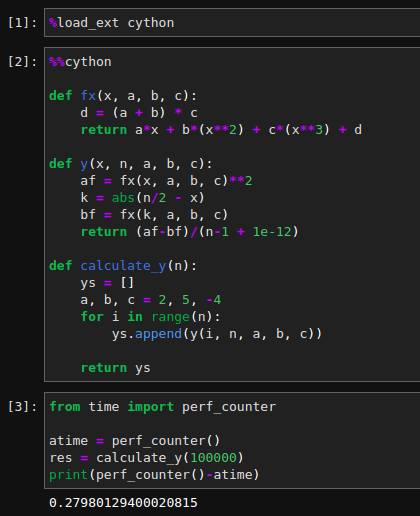
We can show the interactive Cython annotations in Jupyter just like we have generated the HTML file above. To show annotations, pass annotate option to %%cython -a magic command.
Import '.pyx' using pyximport
While developing or debugging, for each change in the '.pyx' file, running setup.py is a repetitive task and cumbersome. Instead, we can dynamically import '.pyx' to Python directly using pyximport without any external build and compilation. pyximport takes care of compiling and building in the background without calling cythonize() internally. So, while importing the '.pyx' file, it will take some time to be imported as a regular Python module.
1import pyximport 2pyximport.install(language_level=3) 3 4from calculate_y_cython import calculate_y_cython 5
Though it is easy to work with Cython using pyximport, there are some limitations with pyximport and it is also not flexible as normal setup.
It is not recommended to use pyximport while distributing Python packages and modules.
Optimize Cython
In the previous section, we have seen in the Cython annotations HTML file that many areas in the code need to be optimized for more speed. There are several ways to improve speed like
- define static types
- use C-libraries and functions
- utilize OpenMP for parallel computing
Define static types to variables
Since Python is a dynamic typing language, we can define C-like data types for Python variables in Cython. To declare C variables, prefix the cdef keyword to the variable declaration which is the same as declaring variables in C.
The syntax for variable declaration is
1cdef type variable_name = initilization_value {optional} 2
The type can be any of the acceptable C data types.
Define function definitions
Python functions are defined using def and C functions in Cython are defined with the keyword cdef. The difference between def and cdef is that with the former declaration, it can be called from anywhere both local and external modules where as cdef functions are only module level. Also, Cython wraps the def function that is defined inside '.pyx' into a Python object. There is another declaration called cpdef which behaves as def when called from outside the module and behaves as cdef inside the module, so it is faster inside the same module function call.
Cython also provides support for writing function definitions just like C. We can define parameter types and return types.
1cdef(or cpdef) type function_name(type parameter1, ...) 2
If no type is specified, the parameters and return values are treated as Python objects which need Python interpretation and they are slow.
Now, make some changes to calculate_y_cython.pyx with static type declarations and function definitions as,
Call the C functions in Cython inplace of Python functions that reduces the Python interaction.
calculate_y_cython.pyx
1ctypedef long int li 2ctypedef long long int lli 3 4cdef lli fx(li x, int a, int b, int c): 5 cdef int d = (a + b) * c 6 return a*x + b*(x**2) + c*(x**3) + d 7 8cdef double y(li x, li n, int a, int b, int c): 9 cdef: 10 lli af = fx(x, a, b, c)**2 11 li k = abs(n//2 - x) 12 lli bf = fx(k, a, b, c) 13 return (af-bf)/(n-1 + 1e-12) 14 15cpdef calculate_y_cython(li n): 16 ys = [] 17 cdef: 18 int a = 2, b = 5, c = -4 19 li i = 0 20 for i in range(n): 21 ys.append(y(i, n, a, b, c)) 22 23 return ys 24
speed can be improved further by disabling bound checking(@cython.boundscheck(False)) and negative indexing(@cython.wraparound(False)) compiler directive instructions.
Check the annotations for the Cython code, most of the yellow lines are now changed to white and only list operations are dark yellow because lists are Python objects. In a later section, we discuss optimizing lists using C arrays.
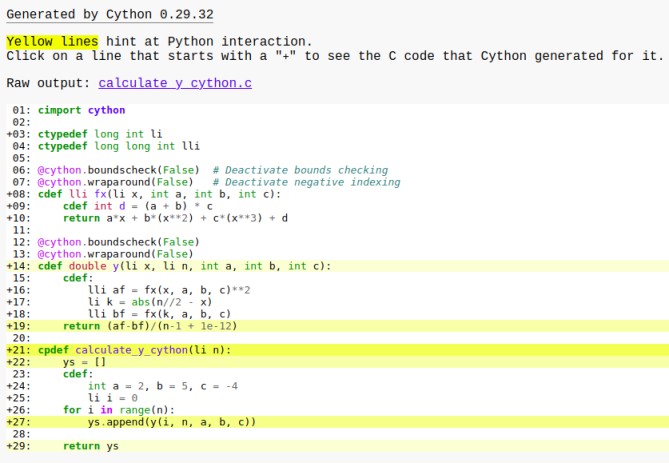
If we check the time for the optimized Cython code,
1%%timeit -n 100 2n = 100000 3res = calculate_y_cython(n) 4 5"""Output: 62.04 ms ± 166 µs per loop (mean ± std. dev. of 7 runs, 100 loops each) 7""" 8
it takes 0.002 seconds which is approximately 100x times faster than the normal Python version that takes 0.19 seconds. By adding only static type and some tweaks, we made Python 100x faster. The powers of Cython are not limited to only static typing. We can further speed up the code with Numpy.
1000x times faster with MemoryView and Numpy
In the Cython annotation snippet above, we see that the list operations of ys are still yellow as list() is a python object and Cython cannot optimize Python objects interaction.
To solve this, we can convert the list type to memoryview arrays, and operations like append() to indexing just like looping in C.
calculate_y_cython.pyx
1cimport cython 2import numpy as np 3cimport numpy as np 4 5ctypedef long int li 6ctypedef long long int lli 7 8@cython.boundscheck(False) # Deactivate bounds checking 9@cython.wraparound(False) # Deactivate negative indexing 10cdef lli fx(li x, int a, int b, int c): 11 cdef int d = (a + b) * c 12 return a*x + b*(x**2) + c*(x**3) + d 13 14@cython.boundscheck(False) 15@cython.wraparound(False) 16cdef double y(li x, li n, int a, int b, int c): 17 cdef: 18 lli af = fx(x, a, b, c)**2 19 li k = abs(n//2 - x) 20 lli bf = fx(k, a, b, c) 21 return (af-bf)/(n-1 + 1e-12) 22 23@cython.boundscheck(False) 24@cython.wraparound(False) 25cpdef calculate_y_cython(li n): 26 cdef double[:] ys = np.empty(n, dtype=np.float64) 27 cdef: 28 int a = 2, b = 5, c = -4 29 li i = 0 30 for i in range(n): 31 ys[i] = y(i, n, a, b, c) 32 return ys 33
Before building the object code, we need to change build instructions to link Numpy as a dependency like following
setup.py
1from distutils.core import setup 2from distutils.extension import Extension 3from Cython.Build import cythonize 4import numpy 5 6ext_modules = [ 7 Extension("calculate_y_function", 8 sources=["calculate_y_cython.pyx"], 9 libraries=["m"], 10 compiler_directives={"language_level": "3"}, 11 ) 12] 13 14setup(name="calculate Y function", 15 ext_modules=cythonize(ext_modules), 16 include_dirs=[numpy.get_include()]) 17
The parameter include_dirs includes external libraries like Numpy here.
1%%timeit -n 1000 2n = 100000 3res = calculate_y_cython(n) 4 5"""Output: 6208 µs ± 11.3 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each) 7""" 8
The fully optimized version of Cython runs at 0.0002 seconds which is approximately 1000x faster than the normal Python code.
The usage of Numpy and MemoryViews in Cython needs separate discussion and I will write about Classes, C-math, Numpy, MemoryViews, and OpenMP in the Advanced Cython series later.
Cython is a very powerful extension that we can use to speed up Python code. Sometimes it may not be possible to convert Python objects straight away like dictionaries (use C++ maps), so it's better to use Cython for repetitive tasks like loops, general functions with simple statements (like declaration and usage only), and math operations.
Learn more about Cython by referencing